Most of the time between starting manuscript and having it accepted after peer-review is spent writing, re-writing and re-arranging content. In Word, keeping track of figure numbers is a big pain, even more so when figures are moved between the main manuscript and the supplement. Moving my Word manuscript to knitr, I first had to decide between RMarkdown and LaTeX. Adding citations and figure references in RMarkdown seems still a bit experimental, and I decided to go with LaTeX.
For my template, I implemented two main features. First, thanks to a tip by Iddo Friedberg, supplemental figures are automatically numbered "S1", "S2", etc. Second, I added a bit of LaTeX magic to interleave parts of the main manuscript and the supplement: If I want to move a paragraph or figure to the supplement, I just wrap it with a "\supplement{ }" command. That is, in the source code, the main and supplemental text are right next to each other, and only in the generated PDF are they separated into two parts of the document.
Of course, fulfilling journal requirements once the manuscript is accepted might still take some time, but compared to the time saved in the previous stages this is quite acceptable.
Here is the template, and here an example PDF generated from the template. (For a complete manuscript, see here.)
Showing posts with label R. Show all posts
Showing posts with label R. Show all posts
Tuesday, June 14, 2016
Tuesday, April 19, 2016
Display element ids for debugging Shiny apps
My current Shiny project contains at least five tables and I constantly forget how they are called. So I whipped up a little bookmarklet that uses jQuery to show the id of each div and input. Some of those can be ignored as they are internal names set by Shiny, but most are the actual names you define in R.
Drag this link to your bookmarks: Show IDs
Drag this link to your bookmarks: Show IDs
Friday, March 11, 2016
New R package: a dictionary with arbitrary keys and values
Coming from Python, the absence of a real dictionary in R has annoyed me for quite some time. Now, I actually needed to use vectors as keys in R:
Under the hood, separate C++ dictionaries (unordered_map) are created for the different types of keys. Using R's flexible types that are reflected in Rcpp (as SEXP), such a dictionary can store both numbers and strings (and other objects) at the same time.
The package is available on GitHub: https://github.com/mkuhn/dict
library(dict)
d <- dict()
d[[1]] <- 42
d[[c(2, 3)]] <- "Hello!"
d[["foo"]] <- "bar"
d[[1]]
d[[c(2, 3)]]
d$get("not here", "default")
d$keys()
d$values()
d$items()
# [[ ]] gives an error for unknown keys
d[["?"]]
Under the hood, separate C++ dictionaries (unordered_map) are created for the different types of keys. Using R's flexible types that are reflected in Rcpp (as SEXP), such a dictionary can store both numbers and strings (and other objects) at the same time.
The package is available on GitHub: https://github.com/mkuhn/dict
Tuesday, March 8, 2016
Avoiding unnecessary memory allocations in R
As a rule, everything I discover in R has already been discussed by Hadley Wickham. In this case, he writes:
Often this won't be the bottleneck, but may be useful at some point.
The reason why the C++ function is faster is subtle, and relates to memory management. The R version needs to create an intermediate vector the same length as y (x - ys), and allocating memory is an expensive operation. The C++ function avoids this overhead because it uses an intermediate scalar.In my case, I want to count the number of items in a vector below a certain threshold. R will allocate a new vector for the result of the comparison, and then sum over that vector. It's possible to speed that up about ten-fold by directly counting in C++:
Often this won't be the bottleneck, but may be useful at some point.
Tuesday, March 10, 2015
Creating composite figures with ggplot2 for reproducible research
So far, I have been preparing composite figures by plotting the data using ggplot2, and then putting the panels together in OmniGraffle or Adobe Illustrator. Of course, every time the data is updated, I would need to go back to the vector editing program. After moving my manuscript from Word to knitr, I figured I should also try to cut out the image editing step.
ggplot2 does not make it easy to put different panels together in a seamless fashion and without any margins. However, by piecing together different StackOverflow answers, I found a way to extract different parts of the figures, and glue them back together with the gtable package. I can now produce a plot like this without a trip to Illustrator!
The solution is still a bit fragile, as the different dimensions of the PNG image and the rows and columns need to be adjusted manually to make it look right. Here is a minimal working example (with some superfluous steps, I'm sure):
The output of the Gist should produce this image:
ggplot2 does not make it easy to put different panels together in a seamless fashion and without any margins. However, by piecing together different StackOverflow answers, I found a way to extract different parts of the figures, and glue them back together with the gtable package. I can now produce a plot like this without a trip to Illustrator!
The solution is still a bit fragile, as the different dimensions of the PNG image and the rows and columns need to be adjusted manually to make it look right. Here is a minimal working example (with some superfluous steps, I'm sure):
The output of the Gist should produce this image:
Monday, September 23, 2013
Introducing parallelRandomForest: faster, leaner, parallelized
Together with other members of Andreas Beyer's research group, I participated in the DREAM 8 toxicogenetics challenge. While the jury is still out on the results, I want to introduce my improvement of the R randomForest package, namely parallelRandomForest.
To cut to the chase, here is a benchmark made with genotype data from the DREAM challenge, using about 1.3 million genomic markers for 620 cell lines in the training set to predict toxicity for one drug (100 trees, mtry=1/3, node size=20):
As you can see, parallelRandomForest is 6 times faster even when not running in parallel, and the memory consumption is about 16 times lower. Importantly, the algorithm is unchanged, i.e. parallelRandomForest produces the same output as randomForest.
For our submission, we wanted to try the simultaneous prediction of drug toxicity for all individuals and drugs. Our hope is that the increased training set enables the Random Forest (RF) to identify, for example, drugs with similar structure that are influenced by similar genotype variations.
It quickly became clear that the standard RF package was ill-suited for this task. The RAM needed by this implementation is several times the size of the actual feature matrix, and there is no built-in support for parallelization. I therefore made several changes and optimizations, leading to reduced memory footprint, reduced run-time and efficient parallelization.
In particular, the major changes are:
The latter two optimizations are especially adapted to the use of RFs on genotype matrices, which usually contain only the values 0, 1, and 2. (Homozygous for major allele, heterozygous, homozygous for minor allele.) The 16-fold reduction in memory consumption seen above is mainly caused by switching to bytes (8-fold reduction) and avoiding extra copies (2-fold reduction).
For our simultaneous mapping strategy, the combined training matrix contains about 120,000 columns and 52,700 rows. Total RAM consumption (including the test set and various accessory objects) was only 18 GB. It took about 2.5 hours to predict 100 trees on 12 CPUs. Both in terms of time and RAM, training with the standard RF package would have been impossible.
The optimizations currently only encompass the functions we needed, namely regression RF. Classification is handled by a separate C/Fortran implementation that I didn't touch. I would estimate that with two weeks of dedicated efforts, all functions of the RF package can be overhauled, and some restrictions (such as forcing the use of bytes) could be loosened (by switching implementations on the fly). However, I do not have the time for this. My question to the community is thus: How to proceed? Leave the package on BitBucket? Ask the maintainers of the standard RF package to back-port my changes? Would someone volunteer for the coding task?
To cut to the chase, here is a benchmark made with genotype data from the DREAM challenge, using about 1.3 million genomic markers for 620 cell lines in the training set to predict toxicity for one drug (100 trees, mtry=1/3, node size=20):
- randomForest (1 CPU): 3:50 hours (230 minutes), 24 GB RAM max.
- parallelRandomForest (1 CPU): 37 minutes, 1.4 GB RAM max.
- parallelRandomForest (8 CPUs): 5 minutes, 1.5 GB RAM max.
It quickly became clear that the standard RF package was ill-suited for this task. The RAM needed by this implementation is several times the size of the actual feature matrix, and there is no built-in support for parallelization. I therefore made several changes and optimizations, leading to reduced memory footprint, reduced run-time and efficient parallelization.
In particular, the major changes are:
- not modifying the feature matrix in any way (by avoiding transformations and extra copies)
- no unnecessary copying of columns
- growing trees in parallel using forked process, thus the feature matrix is stored only once in RAM regardless of the number of threads
- using a single byte (values 0 to 255) per feature, instead of a double floating point number (eight bytes)
- instead of sorting the items in a column when deciding where to split, the new implementation scans the column multiple times, each time collecting items that equal the tested cut-off value
The latter two optimizations are especially adapted to the use of RFs on genotype matrices, which usually contain only the values 0, 1, and 2. (Homozygous for major allele, heterozygous, homozygous for minor allele.) The 16-fold reduction in memory consumption seen above is mainly caused by switching to bytes (8-fold reduction) and avoiding extra copies (2-fold reduction).
For our simultaneous mapping strategy, the combined training matrix contains about 120,000 columns and 52,700 rows. Total RAM consumption (including the test set and various accessory objects) was only 18 GB. It took about 2.5 hours to predict 100 trees on 12 CPUs. Both in terms of time and RAM, training with the standard RF package would have been impossible.
The optimizations currently only encompass the functions we needed, namely regression RF. Classification is handled by a separate C/Fortran implementation that I didn't touch. I would estimate that with two weeks of dedicated efforts, all functions of the RF package can be overhauled, and some restrictions (such as forcing the use of bytes) could be loosened (by switching implementations on the fly). However, I do not have the time for this. My question to the community is thus: How to proceed? Leave the package on BitBucket? Ask the maintainers of the standard RF package to back-port my changes? Would someone volunteer for the coding task?
Monday, April 22, 2013
2D plot with histograms for each dimension (2013 edition)
In 2009, I wrote about a way to show density plots along both dimensions of a plot. When I ran the code again to adapt it to a new project, it didn't work because ggplot2 has become better in the meantime. Below is the updated code. Using the gridExtra package and this hint from the ggplot2 wiki, we get this output:
Source code:
Source code:
Thursday, August 11, 2011
ggplot2: Determining the order in which lines are drawn
In a time series, I want to plot the values of an interesting cluster versus the background. However, if I'm not careful, ggplot will draw the items in an order determined by their name, so background items will obscure the interesting cluster:
One way to solve this is to combine the label and name columns into one column that is used to group the individual lines. In this toy example, the line belonging to group 1 should overlay the other two lines:
 |  |
| Correct: Interesting lines in front of background | Wrong: Background lines obscure interesting lines |
One way to solve this is to combine the label and name columns into one column that is used to group the individual lines. In this toy example, the line belonging to group 1 should overlay the other two lines:
Thursday, March 10, 2011
Comparing two-dimensional data sets in R; take II
David commented on yesterday's post and suggested to put the continuous fitted distribution in the background and the discrete, empirical distribution in the foreground. This looks quite nice, although there's a slight optical illusion that makes the circles look as if they'd be filled with a gradient, even though they're uniformly colored:
 |
| Not-so-good fit |
 |
| Better fit |
Wednesday, March 9, 2011
Comparing two-dimensional data sets in R
I wanted to fit a continuous function to a discrete 2D distribution in R. I managed to do this by using nls, and wanted to display the data. I discovered a nice way to compare the actual data and the fit using ggplot2, where the background is the real data and the circles are the fitted data (the legend is not optimal, but for a slide/figure it's probably easier to fix it in Illustrator):
My data frame includes these columns: x, y, enrichment (the real data), pred (my fitted values).
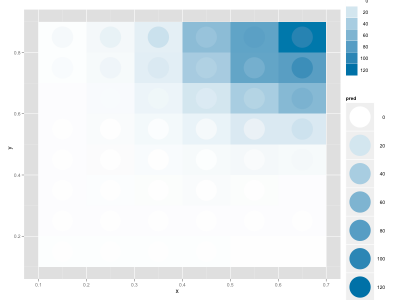 |
| A not-so-good fit |
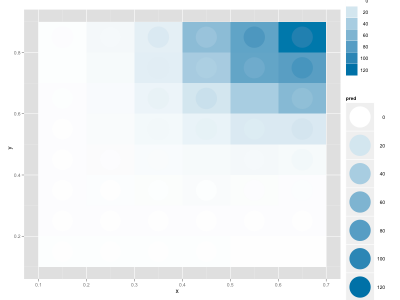 |
| A better fit |
My data frame includes these columns: x, y, enrichment (the real data), pred (my fitted values).
Thursday, September 3, 2009
Learning ggplot2: 2D plot with histograms for each dimension
Update (April 2013): The code below doesn't work anymore with new ggplot2 versions, here is an updated version.
I have two 2D distributions and want to show on a 2D plot how they are related, but I also want to show the histograms (actually, density plots in this case) for each dimension. Thanks to ggplot2 and a Learning R post, I have sort of managed to do what I want to have:
 There are still two problems: The overlapping labels for the bottom-right density axis, and a tiny bit of misalignment between the left side of the graphs on the left. I think that the dot in the labels for the density pushes the plot a tiny bit to the right compared with the 2D plot. Any ideas?
There are still two problems: The overlapping labels for the bottom-right density axis, and a tiny bit of misalignment between the left side of the graphs on the left. I think that the dot in the labels for the density pushes the plot a tiny bit to the right compared with the 2D plot. Any ideas?
Here's the code (strongly based on the afore-linked post on Learning R):
I have two 2D distributions and want to show on a 2D plot how they are related, but I also want to show the histograms (actually, density plots in this case) for each dimension. Thanks to ggplot2 and a Learning R post, I have sort of managed to do what I want to have:
Here's the code (strongly based on the afore-linked post on Learning R):
p <- colour="cyl)<br" data="mtcars," geom="point" hp="" mpg="" qplot="">
p1 <- br="" legend.position="none" opts="" p="">
p2 <- aes="" colour="cyl))<br" ggplot="" group="cyl," mtcars="" x="mpg,">p2 <- br="" fill="NA," p2="" position="dodge" stat_density="">p2 <- axis.title.x="theme_blank()," br="" legend.position="none" opts="" p2=""> axis.text.x=theme_blank())
p3 <- aes="" colour="cyl))<br" ggplot="" group="cyl," mtcars="" x="hp,">p3 <- br="" coord_flip="" fill="NA," p3="" position="dodge" stat_density="">p3 <- axis.title.y="theme_blank()," br="" legend.position="none" opts="" p3=""> axis.text.y=theme_blank())
legend <- br="" keep="legend_box" opts="" p="">
## Plot Layout Setup
Layout <- grid.layout="" ncol="2,<br" nrow="2,"> widths = unit (c(2,1), c("null", "null")),
heights = unit (c(1,2), c("null", "null"))
)
vplayout <- br="" function=""> grid.newpage()
pushViewport(viewport(layout= Layout))
}
subplot <- function="" layout.pos.col="y)<br" layout.pos.row="x," viewport="" x="" y="">
# Plotting
vplayout()
print(p1, vp=subplot(2,1))
print(p2, vp=subplot(1,1))
print(p3, vp=subplot(2,2))
print(legend, vp=subplot(1,2))
Subscribe to:
Posts (Atom)



